Abstract
Introduction Glanzmann's thrombasthenia (GT) is a congenital platelet disorder of autosomal recessive inheritance, affecting ≈1-5:106 individuals (depending on population consanguinity). Patients suffer frequent and relevant bleeding due to a severe platelet aggregation defect. GT is caused by genetic variants in ITGA2B or ITGB3 genes, which encode the αIIbβ3 integrin. DNA sequencing in hundreds of patients by Sanger or short read high-throughput sequencing (HTS) has identified ≈200 causative mutations, including missense, nonsense, and small deletions, insertions or duplications, and splicing variants. Few large gene deletions or rearrangements have been found in rare GT patients by using qRT-PCR or Southern blot approaches. In large series of GT, single heterozygous or no mutations were detected by standard sequencing in about 20% of the cases (Nurden.2015; PMID:25728920). In our series of 40 GT patients from the Spanish Group of Inherited Platelet Defects (GEAPC), Sanger and HTS sequencing was unsuccessful in two unrelated cases.
Objective To explore the value of LongRead Nanopore sequencing technology in genetic characterization of GT cases whose diagnosis was not achieved by conventional ITGA2B and ITGB3 sequencing.
Methods We recruited two males aged 45 (GT-1) and 4 years (GT-2) with mucocutaneous hemorrhagic diathesis (ISTH-BAT 5 and 3) and suspected GT. Platelet phenotyping assays included complete blood count, platelet aggregation (LTA), platelet glycoproteins (GPs), fibrinogen and/or PAC1 binding to αIIbβ3, and platelet secretion of CD62 and CD63 by flow cytometry. DNA (GT-1 and GT-2) and platelet RNA (GT-1 only) was purified. Analysis of ITGB3 and ITGA2B was performed by HTS with a gene panel using an Illumina or Ion Torrent platforms (Bastida.2018,PMID:28983057; Palma-Barqueros.2021,PMID:34516618), and Sanger sequencing. Additionally, DNA spanning regions chr17:46479510-48087045 and chr17:43664909-45074506, which covered ITGB3 and ITGA2B respectively, were sequenced using Nanopore in a MinION device (Oxford Nanopore Technologies), following computational enrichment. Resulting sequences were aligned to the reference genome with Minimap2 and structural variants called with Sniffles.
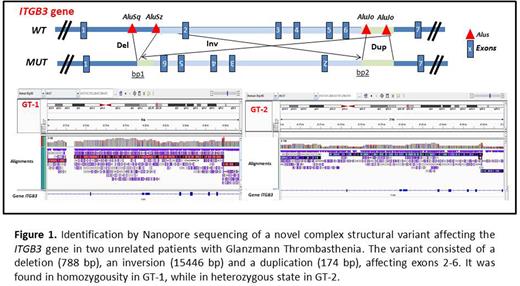
Results Both patients displayed GT type I platelet phenotype: a) normal platelet count and morphology; b) <10% of αIIbβ3 and >90% of GP Ib/IX, GPIa and GPVI; <10% of LTA with arachidonic acid, ADP, epinephrine, collagen or TRAP, and >50% with ristocetin. In GT-1, no ITGB3 cDNA was obtained from platelet RNA, suggesting absent gene expression. Sanger and HTS sequencing of ITGB3 and ITGA2B identified no candidate mutations in GT-1, while in GT-2 the ITGB3 heterozygous splicing variant c.2301+1G>C, inherited from his healthy mother was detected. No mutation was found in his father. Genomic LongRead sequencing by Nanopore revealed in both GT patients the same complex structural variant (SV) in ITGB3, consisting of a deletion (788 bp), an inversion (15446 bp) and a duplication (174 bp), affecting exons 2-6 (Figure 1), which was in homozygosity in GT-1, and in heterozygosity in GT-2. Repetitive Alu elements were identified flanking the breakpoints (Figure 1). Segregation of the SV in the pedigrees was then performed after designing specific PCR primers. In the consanguineous family 1, the mother (the father was deceased), two brothers and two sons of GT-1, all healthy individuals, were heterozygous carriers of SV. Heterozygosity for the SV, inherited from his healthy father, was found in GT-2. Nanopore and standard HTS demonstrated that SV carriers share haplotypes for several common variants in ITGB3, suggesting a founder effect of this complex molecular alteration. No familial relationship or common Spanish geographic origin between the two pedigrees was suspected.
Conclusions DNA analysis by LongRead sequencing using the novel Nanopore technology identified a new ITGB3 complex molecular abnormality shared by two unrelated cases of GT, with no previous definitive molecular defects according to conventional sequencing. These findings support both a replication based mechanism, and a founder effect. Results expand the genetic diversity of GT, and highlight the diagnostic value of longRead sequencing to characterize SV.
Funding:PI20/00926 (ISCIII&Feder);PMP21/00052 (ISCIII&NG EU); GRS2314/A/2021; FMM-AP172142019; SETH Grant for GEAPC
Disclosures
Male:Bayer, Baxalta/Shire/Takeda, Biotest, CSL Behring, Novo Nordisk, SOBI: Consultancy, Research Funding; Bayer, Biotest, CSL Behring, Grifols, Novo Nordisk, Roche, Takeda: Consultancy, Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal